tau P301L
01/3D Structure
? About the 3D Viewer
Mol* (pronounced "molstar") is an open-source molecular visualization tool used by the Protein Data Bank and AlphaFold Database. Learn more at molstar.org.
Controls:
- Rotate: Click and drag
- Zoom: Scroll wheel or pinch
- Pan: Right-click and drag (or two-finger drag)
- Reset: Double-click to reset view
What am I looking at?
This is a predicted 3D structure of the protein. The ribbon diagram shows the protein backbone—helices appear as coils, sheets as arrows, and loops as simple lines. The shape determines how the protein functions: where it binds to other molecules, how it catalyzes reactions, and how mutations might disrupt its activity.
Color legend:
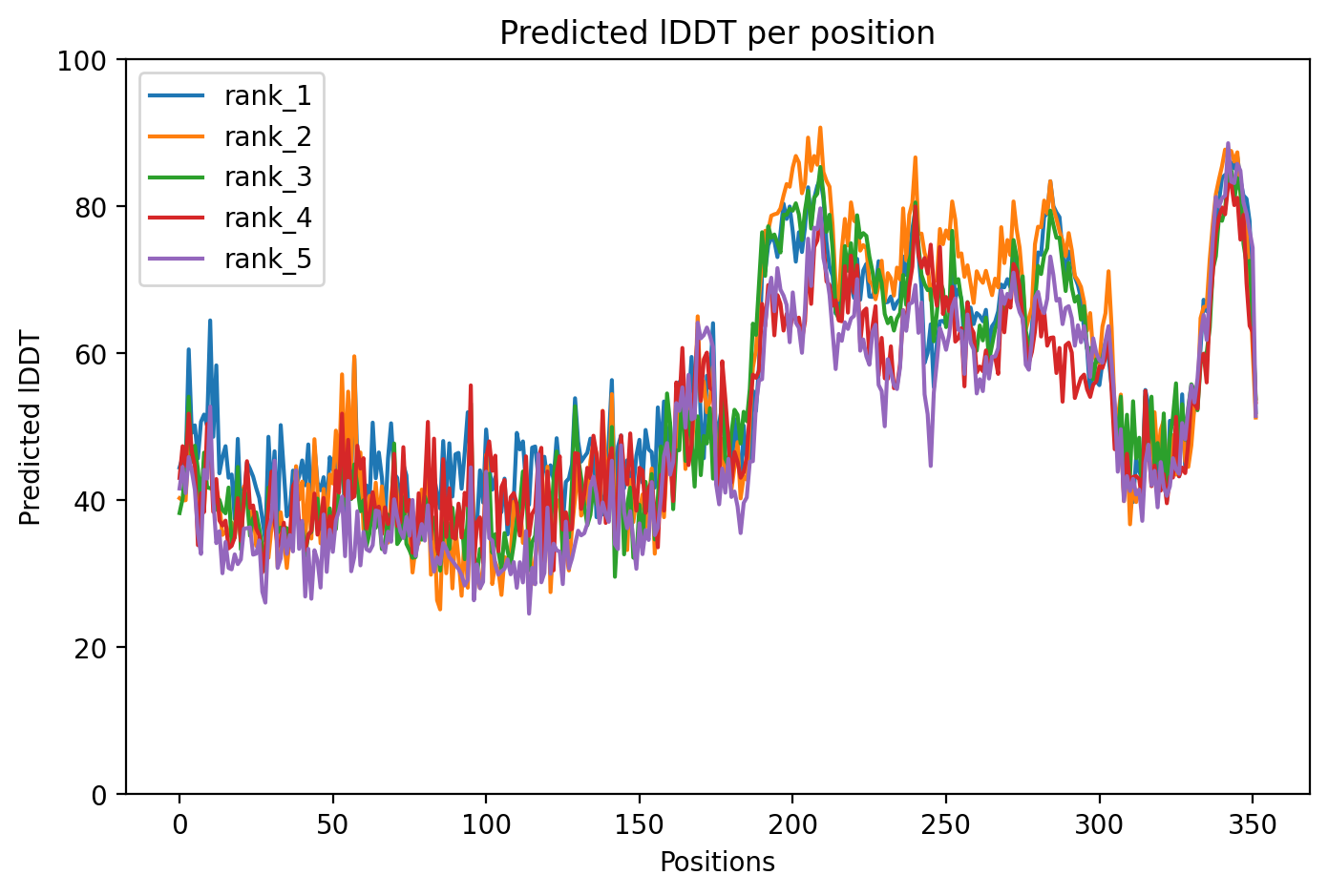
The structure is colored by pLDDT confidence score, which indicates how confident AlphaFold is in each region's predicted position:
- Blue (>90): Very high confidence
- Cyan (70-90): Confident
- Yellow (50-70): Low confidence
- Orange (<50): Very low confidence, likely disordered
02/AI Analysis
TLDR
Tau is a protein that normally helps maintain brain cell structure, but in Alzheimer's disease it becomes damaged and forms toxic tangles. This AlphaFold prediction of the P301L variant (a mutation linked to inherited dementia) shows very low confidence overall, reflecting Tau's naturally floppy, disordered structure that makes it prone to misfolding. The predicted structure reveals that Tau lacks stable 3D shape under normal conditions, which paradoxically enables both its normal function and its dangerous ability to aggregate in disease.
Detailed Analysis
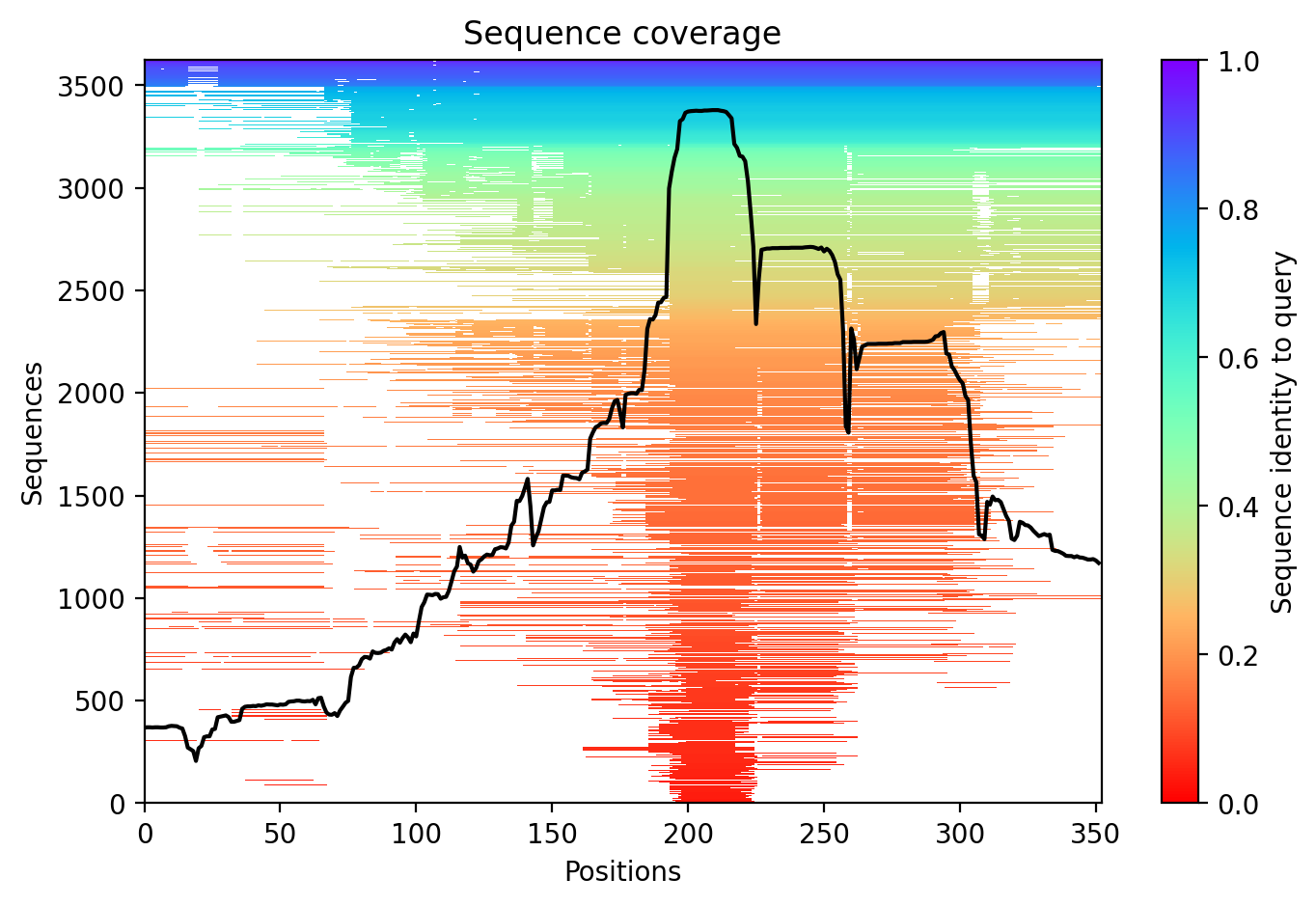
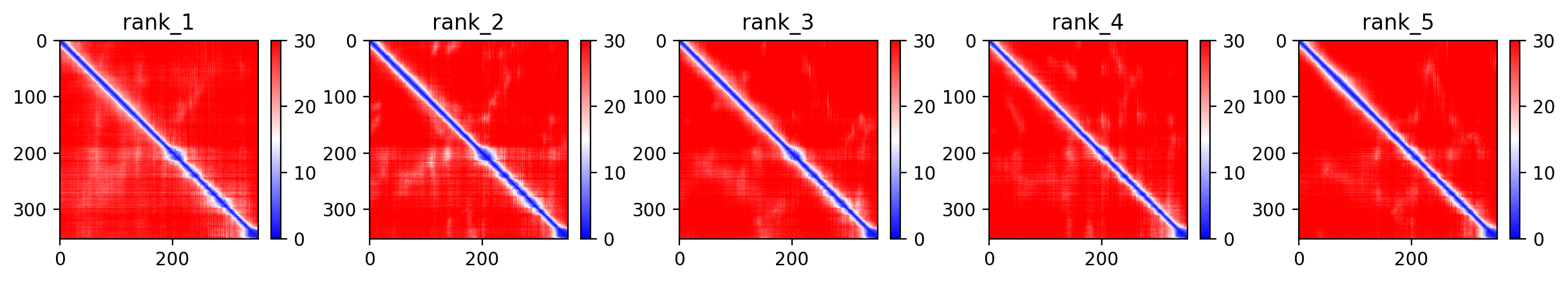
03/AlphaFold Metrics